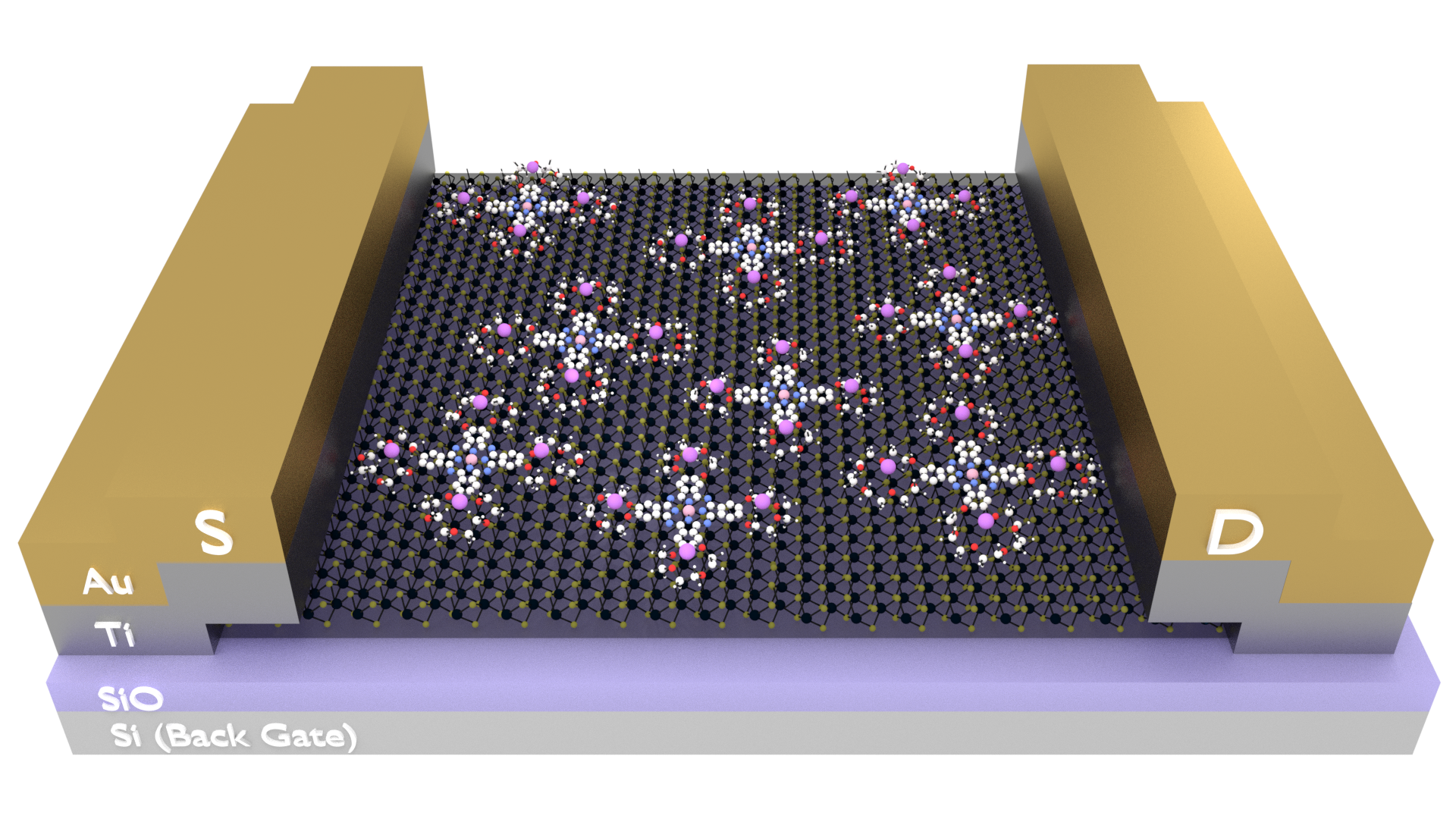
BLENDER MOLECULAR RENDERING
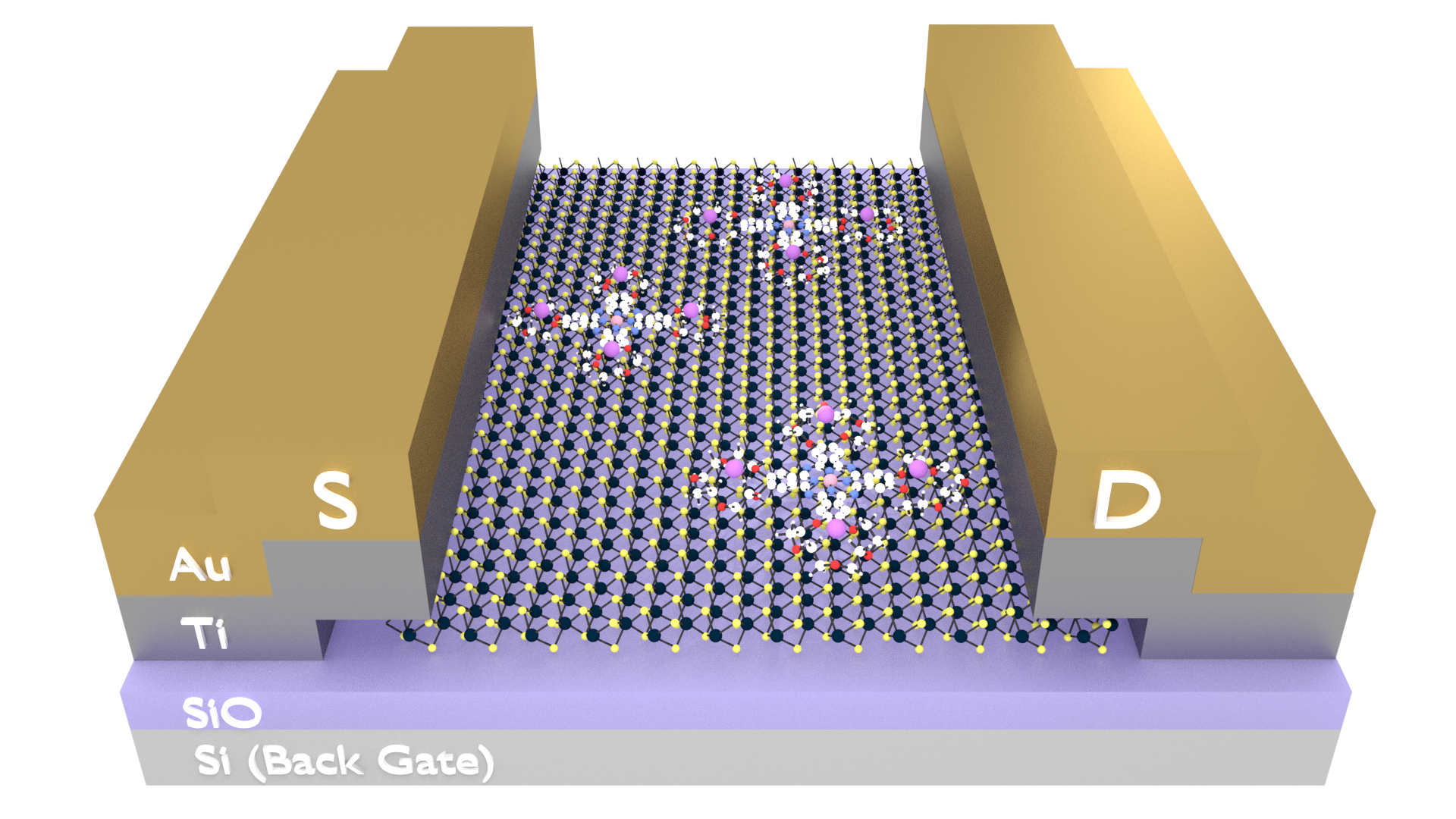
During my research in the Summer of 2017, I was asked to create some images to be used in poster presentations, publications, and a proposal. Most chemistry-based visualization software generates ugly images, but they are preferred because they are spatially accurate and retain the geometries between bonds and atoms. While Blender has a 'prettier' render engine, drawing molecules by hand is both time consuming and inaccurate.
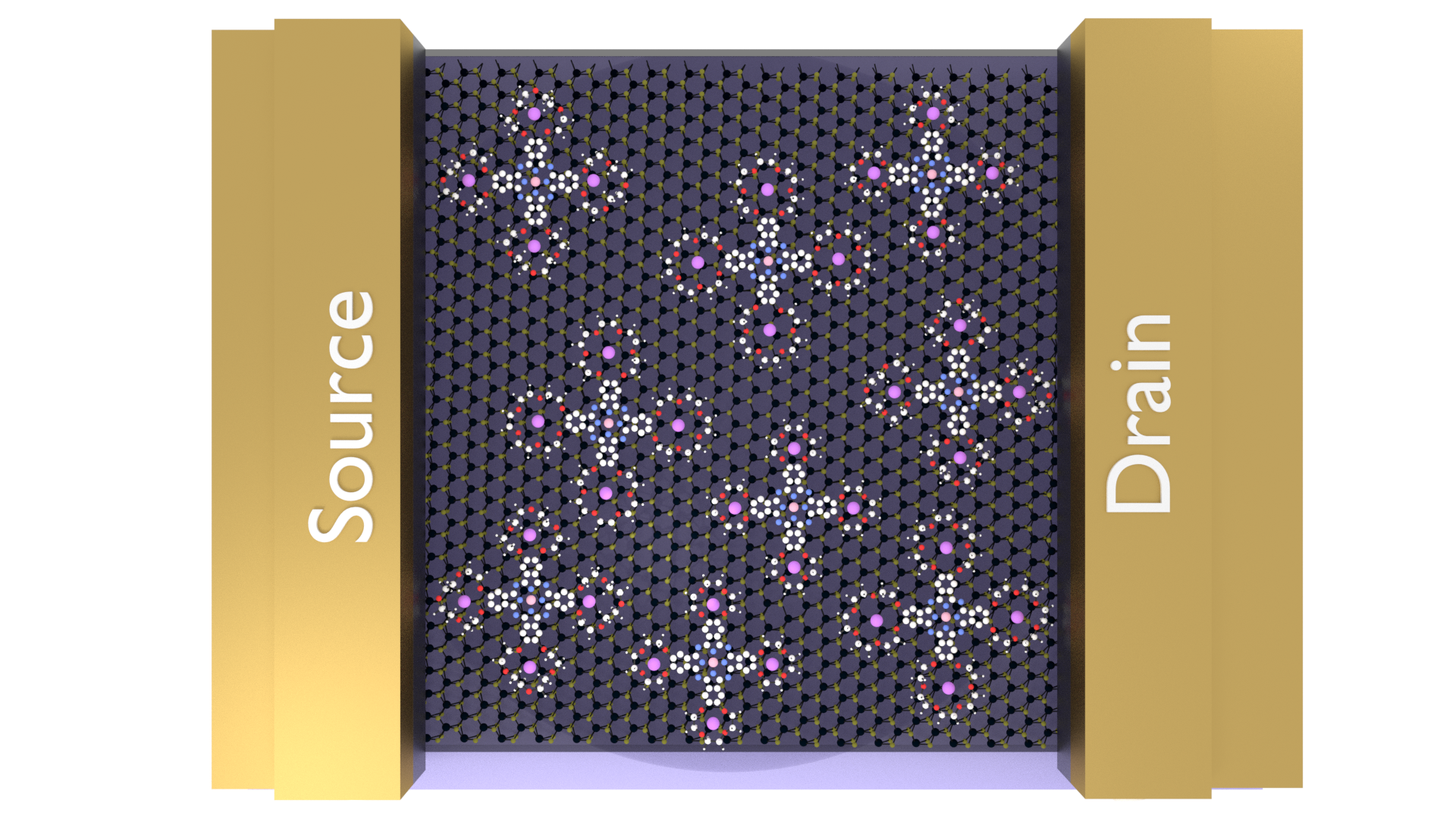
However, using a Python library called PyBel, we are able to convert traditional chemical formats (SMILES, .pdb, and others) into javascript objects, while retaining this geometry. With a little effort, we can teach Blender how to read this file and build the molecule using 3D Blender objects, following spatial and geometric constraints. This is a fantastic little tool that allows the best of both worlds- accuracy of chemistry softwares, with the powerful rendering engine available with Blender.

Because this approach still requires the user to know how to use Python and command prompt entries, it is not very user friendly. Since the Blender API is built in Python, it should be fairly straightforward to convert this program into an addon that users can download run from inside Blender, without any need for external resources. Currently, there are no addons like this at all, making this a project that would be valuable to many other groups looking to make visuals and graphics for publications, etc. Even though I am slow, this is a project I would like to finish.
